Key Findings
>> Blebbistatin and cytochalasin-D inhibit collective migration of 3T3 cells
>> Both drugs impair wound healing in a dose-dependent manner
>> Real-time scratch closure analysis is possible on the Omni live-cell imaging platform
Abstract
The wound healing assay, also known as the scratch assay, is used to understand the role of cell migration in many biological processes. While often used for its low cost and simplicity, variability in scratch creation and quantification methods can influence the results. Here we demonstrate a simple method for consistent scratch creation and automated imaging and analysis using the Omni platform.
Introduction
Cell migration is fundamental for several physiological processes, including (but not limited to) organogenesis, wound healing, and cancer metastasis. It is crucially important to understand the role of cell migration in these processes for potential therapeutic solutions.
Several assays have been developed to elucidate the migratory potential of cells in vitro. Examples include the Boyden transwell chamber, where cells migrate through pores in a membrane; the fence assay, where cells migrate to their non-covered surrounding after removal of a barrier; and the wound healing assay, where a cell-free area is created by wounding a confluent monolayer1. The scratch assay is often used due to its low cost and simplicity2. Multiple methods exist for producing the scratch, with the most common technique being manual wounding of the confluent layer using a pipette tip3.
One of the limitations of manual wounding is the (user-dependent) scratch size inconsistency4. To overcome this, automated methods have been developed to ensure uniform wounding of every sample. An example is the Autoscratch™ Wound Making Tool from BioTek, which creates uniform scratch wounds in 24- or 96-well plates in an aseptic environment, such as a tissue culture hood.
Another limitation of the current scratch assays is the method of data quantification. Generally, a few pictures are taken of the scratch area using a standard benchtop microscope. Especially when many samples need to be analyzed, this is an extremely labor-intensive task resulting in incomplete and inaccurate data.
Automated live-cell imaging and analysis solve these limitations. The Omni is an automated live cell imaging system that can be placed inside a standard cell culture incubator. The entire culture vessel is scanned at set time intervals, providing a complete overview of all the samples without disturbing the cells.
In this study, the Autoscratch™ Wound Making Tool was used to perform a scratch assay in a 96-well plate with confluent layers of NIH-3T3 mouse embryonic fibroblast cells (3T3 cells). As a proof of concept, 3T3 cells were treated with different concentrations and combinations of the cytoskeletal inhibitory drugs blebbistatin and cytochalasin-D.
Materials and Methods
A 96-well plate was seeded with 3T3 cells at a concentration of 20,000 cells per well in Advanced DMEM (Invitrogen) supplemented with 10% Fetal Bovine Serum (FBS; Gibco) and 1% penicillin-streptomycin (PS; Gibco). After 24 h in culture, 96 uniform wounds were created in the cell monolayer using the Autoscratch™ Wound Making Tool. Samples were rinsed with phosphate-buffered saline (PBS; Gibco) to remove detached cells. 100 µl culture medium was added to each well and supplemented with (except for the control) different concentrations of blebbistatin (Bleb; B-0560, Sigma Aldrich; 0.1 - 10 µM), cytochalasin-D (CD; C8273, Sigma Aldrich; 1 - 10 nM) or a combination of both. Thereafter, the well plate was placed on the Omni to obtain high-resolution images at 37°C and 5% CO2 (Figure 1).

The images were automatically uploaded to the Axion Portal, where the Scratch Analysis Module determined the scratch area and migration speed of each sample every hour for a time span of 24 h. The analyzed data was downloaded from the Portal (Figure 2). The scratch area was normalized to t=0 to calculate the wound closure (%) and the average migration speed (µm2/h).

Statistical differences in wound closure were analyzed using two-way ANOVA followed by a Tukey’s multiple comparison test in GraphPad Prism. Differences in migration speed were determined using the one-way ANOVA, followed by a Tukey’s multiple comparison test. Values were considered significant at p<0.05. All data are reported as mean ± standard error of the mean.
Results
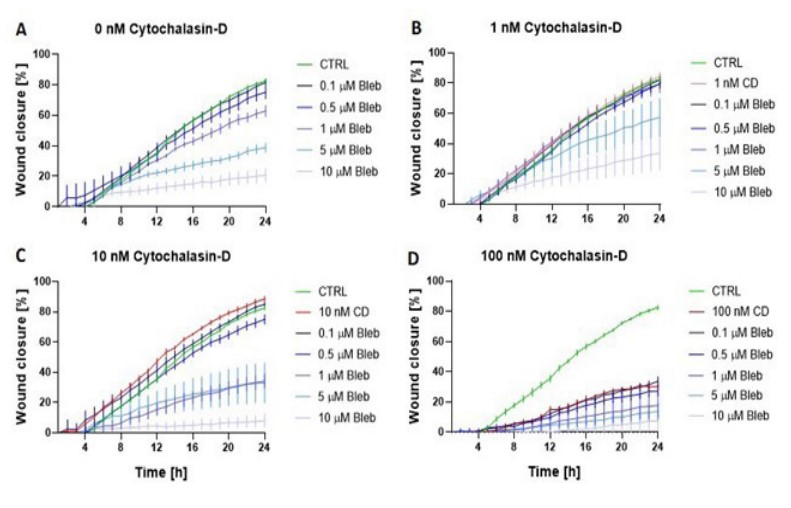
Wound closure was similar in all groups during the first 4 h of the experiment (Figure 3). Wound healing was observed to be compromised over time whenever 5 or 10 µM blebbistatin (Figure 3A, 3B, 3C) was added to the sample. Likewise, the addition of 100 nM of cytochalasin-D had a substantial impact on the wound closure (Figure 3D).
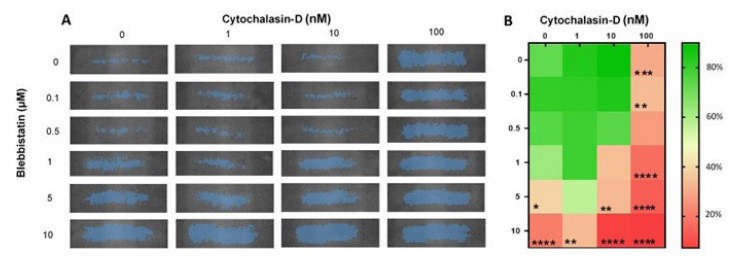
After 24 h, all samples treated with either 10 µM blebbistatin or 100 nM cytochalasin-D had a wound closure under 40% (Figure 4A and 4B). Samples with 10 nM cytochalasin-D had an impaired wound closure when 1, 5, or 10 µM blebbistatin was added (Figure 4B). Although no synergy was detected between these drugs, a trend can be observed that blebbistatin and/or cytochalasin-D impair wound healing in a dose-dependent manner.
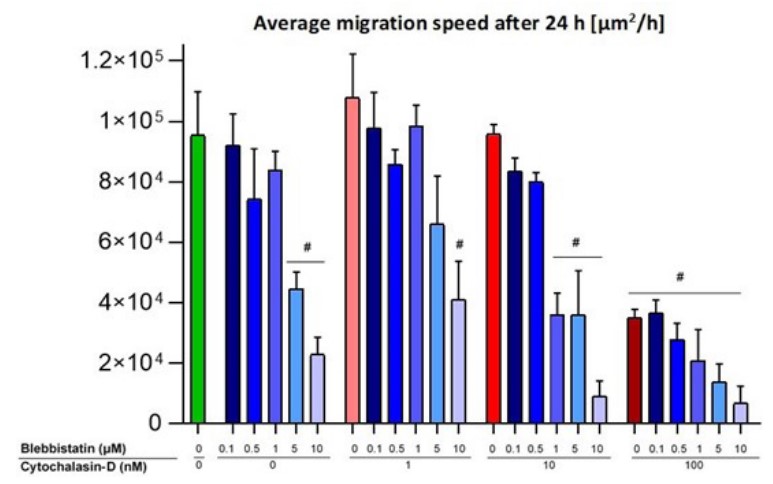
The average migration speed of the control group was 9.5*104 ± 2.8*104 µm2/h (Figure 5). A significantly lower average migration speed could be observed for the 5 µM (4.4*104 ± 1.1*104 µm2/h) and 10 µM (2.3*104 ± 1.1*104 µm2/h) Bleb-treated groups (Figure 5). In addition, the mixed concentrations of 10 µM Bleb + 1 nM CD; 1, 5, and 10 µM Bleb + 10 nM CD; and all samples treated with 100 nM CD had an average migration speed (more than) two times lower than the control group.
Discussion
The wound healing assay is commonly used to understand the migratory potential of cells in vitro. Cell migration is typically studied by creating a wound in a confluent monolayer of cells using a pipette tip. The quality of the results of this assay is a combination of the materials used and the user’s experience, making the assay imprecise and inconsistent. This study solved these problems by using the Autoscratch™ Wound Making Tool to create uniform and consistent scratch areas in every sample.
In addition, gathering kinetic information about wound closure is a tedious and labor-intensive method resulting in shock and possible contamination every time the sample is taken out of the incubator. To overcome this, we used the Omni, an automated live-cell imaging device placed inside the incubator. The Omni scanned the entire well plate and performed automatic image analysis of the wound closure for all 96 wells. Because cellular processes are rarely linear, it is important to use a high temporal resolution, especially for dose- and time-dependent experiments. Here, the time interval was set to 1 hour to understand the short and long-term effects blebbistatin and cytochalasin-D have on 3T3 cells.
Here, it was shown that the cytoskeletal inhibitory drugs blebbistatin and cytochalasin-D had a dose-dependent effect on the wound closure and migration speed of 3T3 cells. Cell migration is driven by the continuous reorganization and turnover of the actin cytoskeleton. This process can be seen as a treadmill, where actin filaments are broken down into monomers at the back of the cell, while at the front, these monomers are used to build up the existing actin filament. When these actin filaments contract with the use of myosin II motors, cellular movement is initiated.
Blebbistatin is a myosin II inhibitor and binds between the nucleotide-binding pocket and the actin-binding site of myosin5,6. Consequently, ATP is not able to initiate tightening in the actin filaments, resulting in an impaired contraction and cell migration mechanism. On the other hand, cytochalasin-D is an inhibitor of the F-actin polymerization process, making it unable to form proper actin filaments7,8. The higher the concentration of blebbistatin, cytochalasin-D, or both, the more wound closure and migration speed were decreased due to inhibition of the actin cytoskeleton.
Conculsion
In this study, we have shown that the Omni easily images a 96-well plate with uniform scratches made by the Autoscratch™ Wound Making Tool. The integrated scratch algorithm accurately determined the scratch area of each well in high-resolution scans that were made every hour for a 24-hour period. We showed that blebbistatin and cytochalasin-D, both cytoskeletal inhibitors, prevent wound closure and decrease the migration speed in a dose-dependent manner.
References
[1.] Riahi, R., Yang, Y., Zhang, D. D. & Wong, P. K. Advances in Wound-Healing Assays for Probing Collective Cell Migration. J. Lab. Autom. 17, 59–65 (2012).
[2.] Kramer, N. et al. In vitro cell migration and invasion assays. Mutation Research - Reviews in Mutation Research 752, 10–24 (2013).
[3.] Ashby, W. J., Zijlstra, A. & Edu, A. Z. Established and Novel Methods of Interrogating Two-Dimensional Cell Migration. Integr. Biol. Quant. Biosci. from nano to macro 4, 1338–1350 (2012).
[4.] Vang Mouritzen, M. & Jenssen, H. Optimized scratch assay for in vitro testing of cell migration with an automated optical camera. J. Vis. Exp. 2018, (2018).
[5.] Kovács, M., Tóth, J., Hetényi, C., Málnási-Csizmadia, A. & Seller, J. R. Mechanism of blebbistatin inhibition of myosin II. J. Biol. Chem. 279, 35557–35563 (2004).
[6.] Caldas, L. A., Seabra, S. H., Attias, M. & de Souza, W. The effect of kinase, actin, myosin and dynamin inhibitors on host cell egress by Toxoplasma gondii. Parasitol. Int. 62, 475–482 (2013).
[7.] Sayyad, W. A., Amin, L., Fabris, P., Ercolini, E. & Torre, V. The role of myosin-II in force generation of DRG filopodia and lamellipodia. Sci. Rep. 5, 1–12 (2015).
[8.] Bashirzadeh, Y., Poole, J., Qian, S. & Maruthamuthu, V. Effect of pharmacological modulation of actin and myosin on collective cell electrotaxis. Bioelectromagnetics 39, 289–298 (2018).
Authors
Lieke Stemkens and Inge Thijssen - van Loosdregt
Axion BioSystems, Atlanta, GA