Key Findings
>> Compared to limiting dilution, DispenCell S3 provided 3x more single cell clones along with impedance-based monoclonality assurance.
>> Using the Omni live-cell imager, monoclonality and clone outgrowth could easily be determined.
>> Impedance and imaging data combined provide double and orthogonal confirmations of the monoclonality of growing clones.
Abstract
Ensuring monoclonality is crucial in generating clonal cell lines for research and production purposes. This application note explores the use of the DispenCell S3, a precise single cell seeder, in combination with the Omni live-cell imager to achieve double, orthogonal monoclonality assurance through both impedance and optical confirmations. The study demonstrates how these two instruments work together to enable reliable single cell cloning, providing clonality verification through controlled seeding and cross-confirmation using both impedance and imaging.
For multiple cell lines, data is provided on clone outgrowth and monoclonality verification, addressing the question whether these two instruments are capable of generating growing clones with double assurance of clonality.
Introduction
Achieving monoclonality in single cell cloning presents significant challenges, particularly when using traditional methods such as limiting dilution.1 This technique involves diluting a cell suspension to a point where individual cells are expected to occupy separate wells in a multi-well plate.1 However, limiting dilution is inherently random, leading to low cloning efficiency. The random distribution of cells can result in multiple cells occupying the same well, while other wells may contain none at all. This significantly reduces the reliability of monoclonality assessments, making it difficult to ensure that the generated cell lines are indeed monoclonal.
This application note introduces the DispenCell S3 single cell seeder and the Omni live-cell imager, devices that can respectively improve the efficiency of single cell seeding and the analysis of monoclonality. Here, we examined their combined effectiveness in improving monoclonality in single cell cloning. CHO and A549 cells were tested as important cellular models.
CHO cells (Chinese Hamster Ovary cells) are widely used in biotherapeutics production due to their ability to perform post-translational modifications and produce complex proteins, making them essential for developing monoclonal antibodies and other biologics.2
The A549 cell line, derived from lung carcinoma, is one of the most extensively studied lung cancer cell lines. Their robustness and ability to divide indefinitely provide valuable insights into cellular mechanisms, drug testing, and lung cancer therapies.3
By using these diverse cell types, the effectiveness of the DispenCell S3 and Omni can be evaluated across various applications, demonstrating their versatility in single cell cloning workflows.
Materials and Methods
Cells and reagents
CHO cells (Cat. CCL-61, ATCC) were used as a model for biotherapeutics production. Cells were maintained in Ham’s F-12 medium (Cat. 11765-054) supplemented with 10% FBS (Cat. 10500-064) and 1% Penicillin-Streptomycin (Cat. 15140-122), all obtained from ThermoFisher.
A549 cells (Cat. P20118, Innoprot) were used as a model for oncology research. The cells were maintained in Ham’s F-12K medium (Cat. 21127-022) with 10% FBS (Cat. 10500-064) and 250 µg/ml Geneticin (Cat. 11811-023), all obtained from ThermoFisher.
Instruments
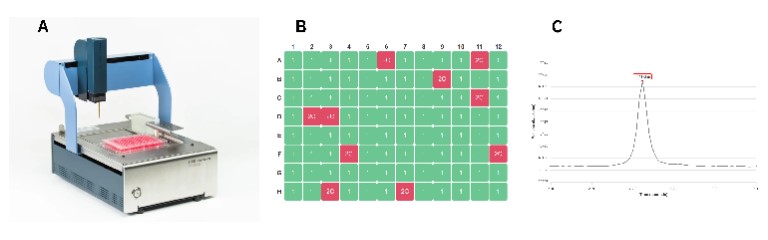
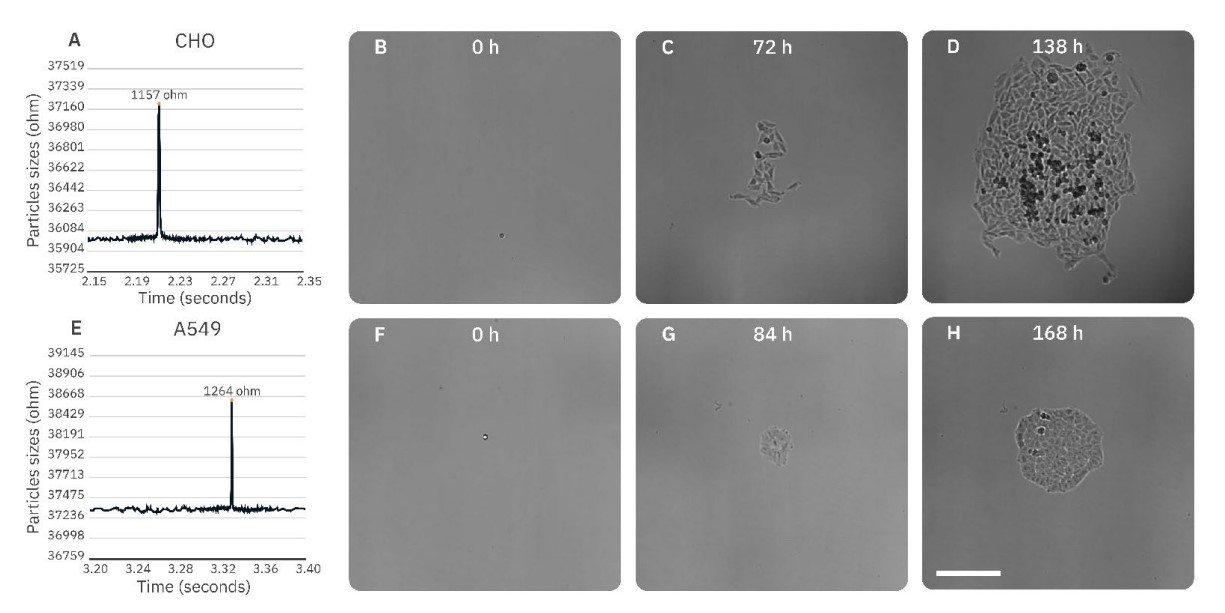
The DispenCell S3 Single Cell Seeder is a highly precise device that seeds single cells based on impedance signals (Figure 1).
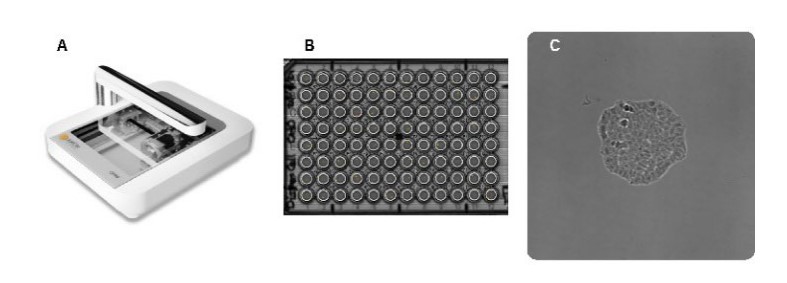
The Omni live-cell imager is a cloud-based live-cell imaging system that enables remote analysis of cell/colony growth and allows users to capture and analyze images from any location (Figure 2).
Workflow Overview
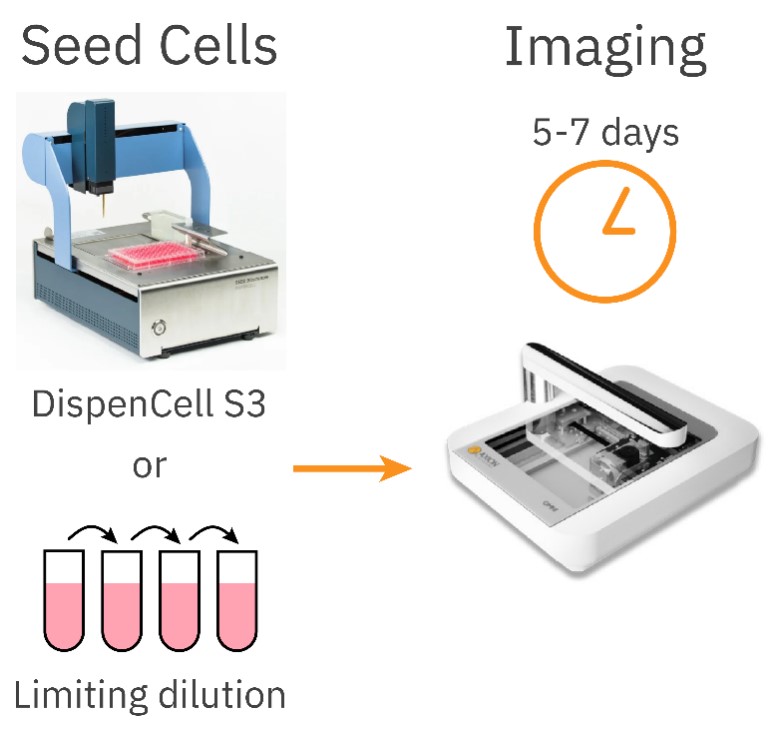
On day 0, single cells were seeded into wells of a 96-well plate using two methods: DispenCell S3 and limiting dilution as a control (Figure 3). Unlike the random nature of the limiting dilution method, DispenCell S3 seeds single cells with much higher efficiency by detecting single cells based on an impedance signal. Each single cell generates a distinct impedance peak, which serves as post-processing proof of clonality. Once seeded, the plates were imaged with the Omni using a 6-hour time interval for 5-7 days.
Single cell seeding with DispenCell S3
Single cells were seeded according to the DispenCell S3 protocol.4 In short, cells were detached and resuspended in culture medium according to the vendor’s protocol. Cells were counted and diluted to a 1.5 x 105 cells/ml suspension. 15 µl of this suspension was added to the DispenMe tube to obtain a final concentration of 1.5 x 104 cells/ml. This tube was placed in the DispenCell S3 to load the sample into the pipette tip. Subsequently, the steps in the DispenCell S3 software were followed to dispense a single cell in each well of a 96-well plate filled with 200 µl of culture medium per well. One well was filled with 20 cells to help with focusing of the Omni.
The impedance profiles obtained with the DispenCell S3 were analyzed to confirm single cell seeding. A particle threshold of 200 Ω was used for both cell types.
The aggregate threshold was set at an impedance of 2000 Ω for the CHO cells and 1700 Ω for the A549 cells.
Limiting dilution control
For the limiting dilution protocol, cells were detached and resuspended in culture medium according to the vendor’s protocol. Cells were counted and diluted to a 1.5 x 105 cells/ml cell suspension. This solution was serially diluted to reach an end concentration of 2.5 cells/ml in 21 ml. All wells of a 96-well plate were filled with 200 µl of this final cell suspension, leading to a seeding density of 0.5 cells/well. With an average of 0.5 cells/well, according to the Poisson probability distribution of the cell count per well, 61% of the wells are empty, 30% of the wells have a single cell, and 9% of the wells have >1 cell. In one well, 5 µl of the 1.5 x 105 cells/ml cell suspension was added to help with focusing of the Omni.
Live-cell imaging using the Omni
After seeding with either the DispenCell S3 or the limiting dilution protocol, the well plate was placed on an Omni inside a cell culture incubator (37 °C, 5% CO2).
The well seeded with a high cell number was used to set the correct focus for the plate, and full brightfield scans were made for 5-7 days with a 6-hour time interval. Images were uploaded to the Axion Portal and analyzed with the integrated Clonogenic Assay Module. If a colony was formed after 5-7 days of culture, images of earlier timepoints were visually analyzed to confirm monoclonality of the colony.
Lastly, the number of wells that obtained a monoclonal colony verified by both the DispenCell S3 and the Omni was determined.
Results
The DispenCell S3 consistently leads to a higher cloning efficiency compared to the limiting dilution method
The DispenCell S3 consistently produced a threefold higher number of colonies compared to the limiting dilution method. This increase is attributed to the higher single cell seeding efficiency of the DispenCell S3 method; seeding more single cells results in more growing colonies. Most importantly, each growing clone generated with the DispenCell S3 method came with solid monoclonality evidence (Figure 4) — a key assurance that the random nature of limiting dilution cannot provide.
For the A549 cells, this resulted in a threefold increase in cloning efficiency with the DispenCell S3 method (75% of the wells formed a monoclonal colony) compared to the limiting dilution method (23%; Figure 5). Likewise, the CHO cells had a threefold increase in cloning efficiency with the DispenCell S3 method (74%) compared to limiting dilution (25%; Figure 5).
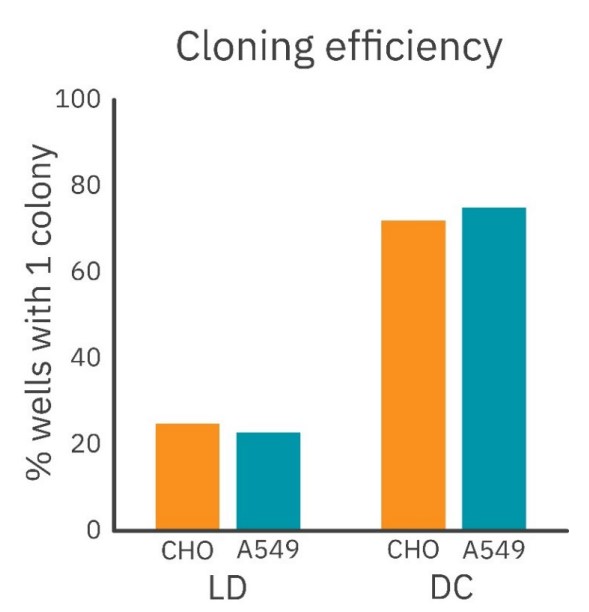
For both cell types, a three-fold increase in monoclonal colony numbers was obtained, underscoring the effectiveness of DispenCell S3 in achieving higher cloning efficiencies.
Double and orthogonal verifications of monoclonality provide a highly robust method
The combination of DispenCell S3 and Omni facilitates double verification of monoclonality with both impedance and imaging, a capability not possible with limiting dilution alone. Compared to other commercial systems offering double optical verifications, this workflow is unique in the sense that it provides orthogonal checks through both impedance and optical confirmations.
For both CHO (64%) and A549 cells (66%) approximately two thirds of the wells were found to contain a monoclonal colony confirmed by both the DispenCell S3 and the Omni. This dual verification enhances the efficiency and reliability of monoclonality assessments.
Conclusion
In summary, this study highlights the advantages of using DispenCell S3 in combination with the Omni for achieving double and orthogonal assurance of clonality through both impedance and optical confirmations in single cell cloning workflows. The increased colony numbers and enhanced verification methods provide compelling evidence for this approach, potentially revolutionizing the generation of clonal cell lines and benefiting both research and industry with simple, compact and affordable instruments.
References
1. Gross A., Schoendube J., Zimmermann S., Steeb M., Zengerle R., and Koltay P. (2015) Technologies for Single-Cell Isolation. Int. J. Mol. Sci. 16(8) 16897-16979. https://doi.org/10.3390/ijms160816897
2. Fischer S., Handrick R., and Otte K., The art of CHO cell engineering: A comprehensive retrospect and future perspectives, Biotech. Adv. (2015) 10.1016/j.biotechadv.2015.10.015
3. Korrodi-Gregório L., Soto-Cerrato V., Vitorino R., Fardilha M., Pérez-Tomás R. (2016) From Proteomic Analysis to Potential Therapeutic Targets: Functional Profile of Two Lung Cancer Cell Lines, A549 and SW900, Widely Studied in Pre-Clinical Research. PLoS ONE 11(11): e0165973. https://doi.org/10.1371/journal.pone.0165973
4. DispenCell S3 User Manual, SEED Biosciences SA, Epalinges, Switzerland, 2024
Authors
Inge Thijssen – van Loosdregt, Staff Application Scientist
Axion Biosystems
Arnaud Gelb, Application Scientist
Georges Muller, CEO
SEED Biosciences