Key Findings
>> Large numbers of embryoid bodies can be tracked and analyzed with the Omni
>> Whole-well imaging ensures consistent measurements across time
>> Metrics such as area, diameter, and roundness can offer insight into culture health and quality
>> Upstream quality control of embryoid bodies is important to improve downstream differentiation protocols
Abstract
Embryoid bodies (EBs) are three-dimensional (3D) aggregates formed from pluripotent stem cells (PSCs) and are often used as an intermediate step between two-dimensional (2D) stem cell cultures and in vitro differentiation of PSCs into various cell types. Overall population homogeneity in size, shape, and number of EBs is critical for successful differentiation protocols and to obtain high yields of differentiated cell types. To improve and streamline the embryoid body workflow prior to in vitro differentiation, we present an automated method to track and analyze organoids using the Omni platform.
Introduction
Embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs) can differentiate into all three germ layers – endoderm, mesoderm, and ectoderm. Pluripotent stem cell aggregates, known as embryoid bodies, emulate aspects of early morphogenesis and are often used as an intermediate step during in vitro differentiation into more specialized cell types. Past studies have demonstrated the formation of complex structures during EB differentiation, including tissues resembling the brain1, optic-cup2, thyroid3, and many more4. Although EBs allow for the lineage-specific differentiation of PSCs, differentiation efficiency and production yields are highly dependent on size distribution and homogeneity of the starting population. The handling of EBs can cause variation between batches and influence the outcome of differentiation protocols. For instance, smaller EBs may not survive the harsh conditions of some differentiation protocols, while larger EBs can develop necrotic cores5. Furthermore, EB populations of varying sizes have a direct impact on the yield of terminally differentiated cells.
The Omni live-cell imaging platform can image cultures directly from the incubator. With the Organoid Analysis Module, it can identify, track, and analyze large numbers of embryoid bodies across an array of culture vessels. Using whole-well brightfield imaging, it can analyze embryoid body populations for area, diameter, roundness, and count (Fig. 1B). In contrast to current methods that rely on manually acquiring images from a standard microscope followed by analysis with third-party software, the Omni allows for easy, automated characterization of embryoid bodies prior to the initiation of differentiation. By facilitating upstream quality control, the Omni greatly reduces the amount of time and effort the user must spend in optimizing long-term differentiation protocols. Furthermore, the Omni can provide guidance in identifying key morphological features that are needed for successful differentiation, improving the final yield and reducing overall culture costs. Here, we present a workflow to monitor iPSC-derived EB cultures using the Omni platform (Fig. 1A).
Materials and Methods
Cells and reagents
Human induced pluripotent stem cells (iPSCs; Cat. 200-0511) were obtained from Stem Cell Technologies (Vancouver, CA). Human iPSC media was composed of mTesR Plus (Stem Cell Technologies, Cat.100-0276). ESC-qualified Matrigel (Cat. 354277) was obtained from Corning (Corning, NY).
iPSC culture
Human iPSCs were thawed and cultured according to the supplier recommendations. Briefly, one vial of iPSCs was thawed and plated at different densities on Matrigel-coated 6-well plates. On Day 6-8 post thaw, the well with optimal iPSC colony density for passaging was selected. ReLeSR (Stem Cell Technologies, Cat. 05872) was used to passage iPSCs from the optimal well and cells were split at a ratio of 1:30 into a new Matrigel-coated 6-well plate. Medium changes were performed every other day to maintain iPSC cultures.
Embryoid body (EB) formation
First, the AggreWellTM 800 24-well plate (Stem Cell Technologies, Cat. 34811) was prepared by adding Anti-Adherence Rinsing Solution (Stem Cell Technologies, Cat. 07010) to each well being used. The plate was centrifuged at 1300 x g, followed by aspiration of the Anti-Adherence rinsing solution. Seeding medium was prepared by adding 5 µL of Y-27632 (Stem Cell Technologies, Cat. 72302) to 2.5 mL of mTeSR Plus media and then 1 mL was added to the same well that was treated with the Anti Adherence Rinsing Solution.
iPSCs were passaged using Gentle Dissociation Reagent (Stem Cell Technologies, Cat. 100-0485) by adding 3 mL to one well of the 6-well plate and incubating at 37°C for 8 minutes. A 1 mL pipettor was used to gently resuspend the cells and transferred to a 15 mL conical tube. Viable cells were stained with trypan blue and counted using a hemocytometer. Next, dissociated iPSCs were centrifuged and resuspended in 1.5 mL of seeding medium to obtain a final concentration of 150,000 cells/mL.
One mL of cell suspension was added to the treated well of the AggreWellTM 800 24-well plate that contained 1 mL of seeding medium to bring the final concentration to 500 cells/microwell. The plate was centrifuged at 300 x g for 5 minutes to capture the cells in the microwells. Three days post seeding, EBs were removed with a wide bore 1 mL pipette tip and transferred to a 6-Well Ultra-Low Adherent Plate (Stem Cell Technologies, Cat. 27145).
Omni imaging platform
The Omni platform was used to measure growth of EB cultures over time inside a cell culture incubator. During brightfield acquisition, the camera scans the sample stage and acquires a series of sequential images. These are stitched together to form an image of the total surface area of the culture vessel. All images are uploaded to the Axion Portal where they can be analyzed using AxIS Vue. For EB monitoring, the Organoid Analysis Module was used for data analysis (Fig. 1). Between scans, EB plates were placed on the orbital shaker for long-term culture.
Results
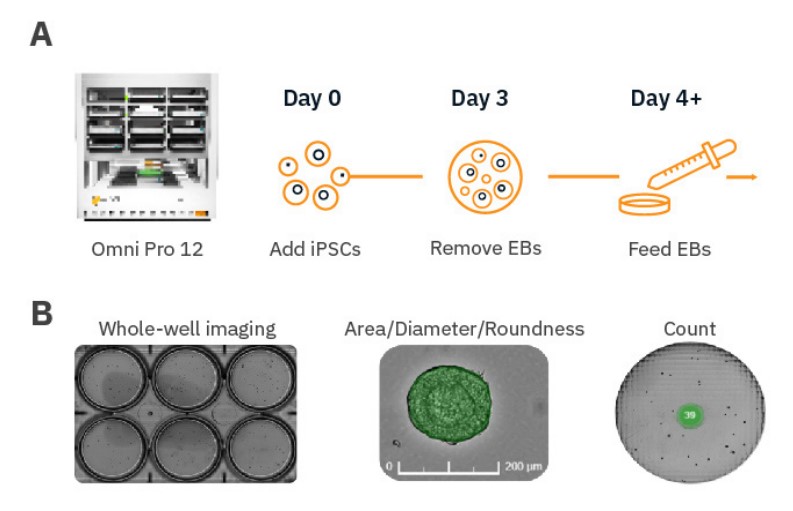
Whole-well imaging ensures counting of all embryoid bodies
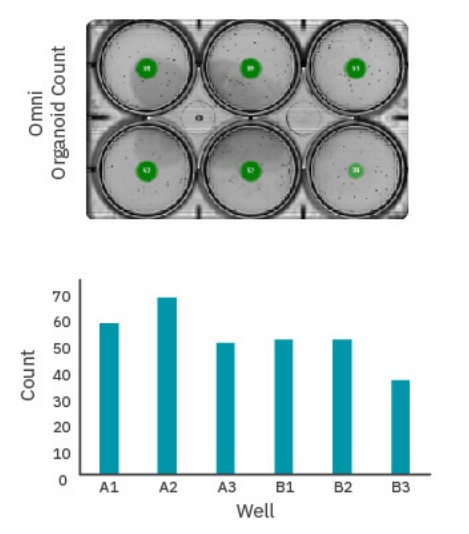
Too many EBs in a culture vessel can result in early fusion of EBs, resulting in loss of overall yield and an increase in the size distribution over time. Maintaining a specific number of EBs in a culture vessel can be critical to ensuring homogenous populations. The dorsal forebrain organoid differentiation protocol (Stem Cell Technologies), for example, recommends a maximum of 40 EBs per well10. While manual counting with conventional microscopy can be time-consuming, Omni’s automated counting simplifies the process.
For EBs cultured on an orbital shaker, distribution can vary between timepoints at any single imaging site. The Omni’s whole-vessel scans ensure each EB is always counted (Fig. 2).
Embryoid body morphology can be characterized in real-time
Optimal EB size is critical in many differentiation protocols. For example, efficient differentiation of EBs into cardiomyocytes is highly dependent on EB size6-8. In contrast to spontaneous EB formation, common procedures such as forced aggregation techniques (e.g., AggrewellTM plates), hanging drop, and microcontact printing have reduced EB size variability but still require monitoring of EB growth over time to determine the optimal size for differentiation. The Omni can analyze embryoid body size, including diameter.
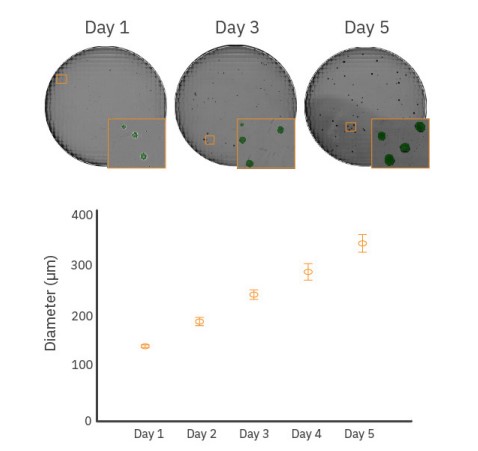
As shown in Figure 3, whole-well scanning was performed every day for 5 days to monitor EB diameter and the acquired images were analyzed to determine the average diameter for each timepoint. EB diameter increased from approximately 150 µm on Day 1 to 350 µm by Day 5. Additionally, EB roundness can be an indicator of the EB state and overall homogeneity of the EB cultures. Roundness values of 0.85 and higher indicate very round EBs. A decrease in EB roundness may be due to changes in EB composition from spontaneous or directed differentiation9.
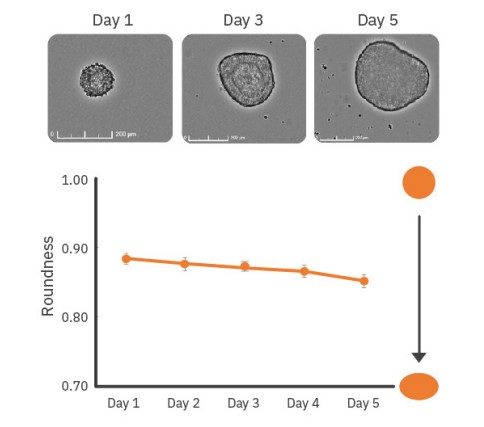
As shown in Figure 4, EBs monitored over 5 days showed a small decline in roundness over time, potentially indicating differentiation or formation of more complex structures within the EBs. Minimal changes in roundness over time are expected for most stable cultures. Large or drastic changes in roundness (r < 0.5) may be an indicator of unhealthy cultures (e.g., EB dissociation).
Conclusion
In this study, we demonstrate an automated approach for EB monitoring to streamline the process of tracking EB size, shape, and count in order to yield consistent and high-quality cultures for downstream differentiation protocols. The whole-well imaging capabilities of the Omni platform are ideal for optimizing stem cell workflows.
References
[1.] Trujillo et al. Complex Oscillatory Waves Emerging from Cortical Organoids Model Early Human Brain Network
[2.] Development. Human brain organoids assemble functionally integrated bilateral optic vesicles. Cell Stem Cell 25, 558–569, October 3, (2019).
[3.] Gabriel et al. Human brain organoids assemble functionally integrated bilateral optic vesicles. Cell Stem Cell 28, 1740–1757 October 7, (2021).
[4.] Longmire, T. A. et al. Efficient derivation of purified lung and thyroid progenitors from embryonic stem cells. Cell Stem Cell 10, 398–411 (2012).
[5.] Kim, J. et al. A. Human organoids: model systems for human biology and medicine. Nat. Rev. Mol. Cell Biol. 21, 571–584 (2020).
[6.] Pettinato, G., Wen, X. & Zhang, N. Formation of Well-defined Embryoid Bodies from Dissociated Human Induced Pluripotent Stem Cells using Microfabricated Cell-repellent Microwell Arrays. Sci Rep 4, 7402, (2014).
[7.] Burridge, PW et al. Improved Human Embryonic Stem Cell Embryoid Body Homogeneity and Cardiomyocyte Differentiation from a Novel V-96 Plate Aggregation System Highlights Interline Variability, Stem Cells, 25, 4, 929–938 (2007).
[8.] Mohr JC et al. The microwell control of embryoid body size in order to regulate cardiac differentiation of human embryonic stem cells. Biomaterials, 7:1885-93. (2010).
[9.] Hwang, YS et al. Microwell-mediated control of embryoid body size regulates embryonic stem cell fate via differential expression of WNT5a and WNT11 106, 40, 16978-16983 (2009).
[10.] Satthenapalli, R, et al. Stage-specific regulation of signaling pathways to differentiate pluripotent stem cells to cardiomyocytes with ventricular lineage. Stem Cell Res Ther 13, 185 (2022).
[11.] STEMdiff™ Dorsal and Ventral Forebrain Organoid Differentiation Kits. https://www.stemcell.com/products/ stemdiff-dorsalforebrain-organoid-differentiation-kit.html
Authors
Denise Sullivan & Daniel Millard
Axion BioSystems, Atlanta, GA

